Author: Waghmare Sandesh Tulshiram
1. Functional Genomics
Functional genomics refers to the development and application of global (genome-wide or system-wide) experimental approaches to assess gene function by making use of the information and reagents provided by structural genomics. It is characterized by high-throughput or large-scale experimental methodologies combined with statistical or computational analysis of the results. Functional genomics as a means of assessing phenotype differs from more classical approaches primarily with respect to the scale and automation of biological investigations. A classical investigation of gene expression examines how the expression of a single gene varies with the development of an organism In Vivo. Gene function is inferred from the resulting phenotype when the particular gene is mutated. Genomics is changing the way genetics by performing Global, high-throughput approaches. Genomics approaches are being applied to both forward and reverse genetics
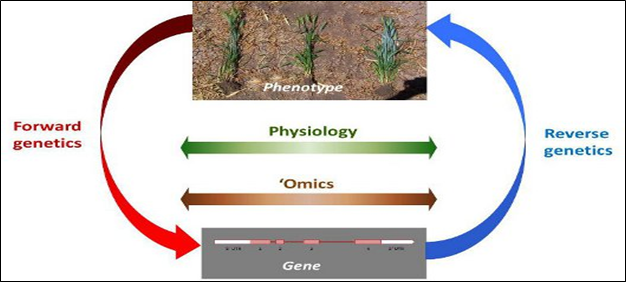
Fig.1: Forward and reverse genetics
1.1. Forward genetics
Forward genetics encompasses several means of identifying the genotype that is responsible for a phenotype. Forward genetics starts with identification of interesting mutant phenotype. Forward genetics seeks to find the genetic basis of a phenotype or trait. Automated DNA sequencing generates large volumes of genomic sequence data relatively rapidly. The main objective of forward genetics is to discover the function of genes defective in mutants. Genomics approaches to forward genetics
- Insertional mutagenesis
- Genetic mapping
- Expression analysis using Microarray
- Candidate gene approach
- Exmore sequencing Insertional mutagenesis is mutagenesis of DNA by the insertion of one or more bases. Insertional mutations can occur naturally, mediated by virus or transposon, or can be artificially created for research purposes in the lab 1.1.1.Genetic mapping
1.1.2. Expression analysis using Microarray
Microarray is a very promising technology for identification of genes in transcription deficient mutants. The approach of using phenotypically similar mutants minimizes the number of candidate genes for sequencing, due to the reduction of genes which are secondarily affected by the mutation. Both cDNA and Affymetrix microarray platforms successfully pinpoint the gene which is up-regulated or down regulated due to the induced or natural mutation events.
1.1.3. Candidate gene approach
This approach is appropriate for plants where mutant collections, represented by multiple independent mutant alleles are available. The major difficulty with this approach is that in order to choose a potential candidate gene for the mutation. Then this gene can be a potential candidate for the mutation in the investigated plant and the principle proof that this candidate gene is responsible for the observed phenotype is coming from comparative sequence analysis of all available mutant alleles in the particular locus.
1.1.4. Exome sequencing
Exome sequencing is a influential method to selectively sequence the coding regions of the genome. This method can be combined with target-enrichment strategies, which give possibility to selectively capture genomic regions of interest from a DNA sample prior to sequencing.
1.2. Reverse genetics
Reverse genetics seeks to find what phenotypes arise as a result of particular genetic sequences. Reverse genetics starts with a known gene and alters its function by transgenic technology. Reverse genetics however, due to the advent of advances in sequencing is employed to seek out what sequences underlies the phenotype. Reverse genetics starts with known genes e.g., from genomic sequencing. Major goal of reverse genetics is to determine function through targeted modulation of gene activity
Genomics approaches to reverse genetics
- Gene silenencing
- TILLING (Targeting Induced Local Lesions in Genomes)
Gene silencing using double stranded RNA is known as RNA interference (RNAi), and the development of gene knockdown using Morpholino oligos, have made disturbing gene expression. RNAi creates a specific knockout effect without actually mutating the DNA of interest. RNAi acts by directing cellular systems to degrade target messenger RNA (mRNA).
1.2.2. TILLING (Targeting Induced Local Lesions in Genomes)
TILLING is based on the use of a mismatch-specific endonuclease ( CelI), which finds mutations in a target gene containing a heteroduplex formation. The technique involves PCR amplification of the target gene using fluorescently labeled primers, formation of DNA heteroduplex between wild type and mutant alleles (PCR products, corresponding to the mutant and wild type alleles are heated and then slowly cooled), followed by endonuclease digestion specifically cleaving at the site of an EMS induced mismatch. One of the greatest benefits of the TILLING approach is that it does not involve genetic manipulations, that results in Genetically Modified Organisms (GMO), which are not legal for agricultural applications in many countries In EcoTILLING genomic DNA sequence is used to identify the mutations created using endonuclease (CEL I). These techniques are independent of genome size, ploidy level and reproductive system of plants. They can be applied not only to model organisms but also to economically important crops.
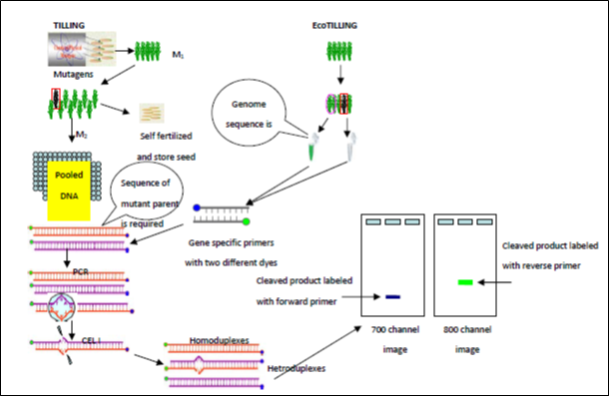
Fig.2: Schematic representation of TILLING and Eco-TILLING
References
Muhammad R., Guangyuan H., Yang G. and Ziaf K. 2011. Relevance of tilling in plant genomics. Australian j. crop sci. A 5(4):411-420
Eva, M. Y., Moresco , Xiaohong L., and Bruce, B.2013. Recent Technological Advances and Forward Genetics in Mice. The American J. of Pathl.182(5):1463-1473
About Author / Additional Info:
Assistant professor. K.K. Wagh College of Agricultural Biotechnology, Nashik