Authors: Devendra Singh1 and Neelam Geat2
1Ph.D. Scholar, Division of Microbiology, IARI, New Delhi, 110012
2Ph.D Scholar, Division of Plant Pathology, IARI, New Delhi, 110012
Typing
Identifying different types of organisms within a species is called typing. In bacterial strain typing, both phenotyping and genotyping can be used.
1. Phenotyping: The most common phenotyping methods are:-
• Serotyping
• Resistotyping
• Phagetyping
• Bacteriocin typing
• Biotyping
Disadvantage of these commonly applied technique may be low discriminatory power. In recent year the quality of microbiological assays has been increased by using molecular biological technique as-
a. FAME analysis: It is very successful because fatty acids are the major constituents of lipid and lipopolysaccharide in microbial cells and have been extensively used for taxonomic purposes.
b. SDS-PAGE and MALDI-TOF- Typing of extracted cell protein.
2. Genotyping methods:
A. Plamid profile analysis
B. Chromosomal DNA analysis
C. Pulse field gel electrophoresis (PFGE)
D. DNA probes in detection and typing of micro-organism
E. Ribotyping
F. PCR (Polymerase Chain Reaction)
a) Real time PCR (RT-PCR)
b) Reverse Transcription PCR
c) Multiplex PCR
d) Repetitive sequence based PCR (Rep-PCR)
G. RFLP (Restriction Fragment Length Polymorphism)
H. RAPD (Random Amplified Polymorphic DNA)
I. AFLP (Amplified Fragment Length Polymorphism)
J. MLST (Multilocus Sequence Typing)
K. Analysis of repetitive DNA sequences
L. MLVA (Multilocus variable Number of Tandom Repeats Analysis)
M. DNA microarray
N. DGGE (Denaturing Gradient Gel Electrophoresis)
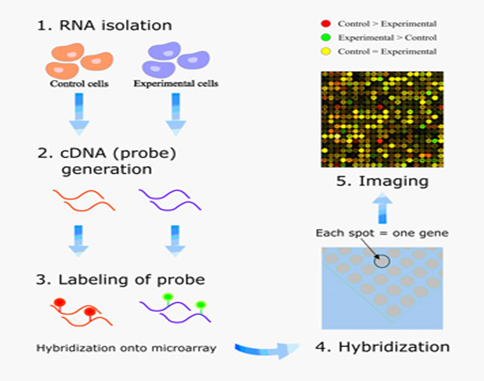
Detail description of DNA Microarray:
A. A microarray is a collection of DNA probes attached in an ordered pattern onto a solid surface. These probes can be used to detect the presence of complementary sequences in bacterial isolates, and thus can detect marker genes in specific bacterial strains.
B. Microarrays combine the potential of simultaneous bacterial identification and speciation. This method is versatile and make it possible to detect and discriminate different bacterial samples on a single slide
C. DNA microarray based approach is used for quick detection and identification of bacteria using species-specific oligonucleotide probes designed for specific regions of various targated genes.
D. Gene expression profile analysis technique is called DNA microarray or DNA chip.
Microarray procedure:
• Isolation of mRNA from a given sample. In practice, mRNA is present in too low amount for direct use. Consequently, the mRNA sequences must first be amplified. Reverse transcriptase (RT) is used to generate cDNA from the mRNA. The cDNA may then be amplified by PCR. (These two steps constitute the RT-PCR procedure).
• Its first chain may be marked with fluorescent dye and denatured to a single stranded DNA.
• At last, the denatured cDNA or denatured marked cDNA is applied to the array. Hybridization will take place. In this way, based on the hybrid, which reveals the presence of mRNA, information about gene expression is generated.
Applications:
i. Microarray comprised of several thousand probes on glass slides so that global gene expression can be measured in a single experiment.
ii. Microarray can be used to identify microorganisms. Such approach can be used to differentiate between closely related strains by differences in their hybridization patterns. For example- identification chips (ID) have been used in the food industry to detect particular pathogenic strain like E.Coli 0157:H7.
iii. The array are large enough and dense enough that the transcription pattern of an entire genome (the transcriptome) can be analysed.
About Author / Additional Info:
I am pursuing Ph.D in Microbiology from IARI, New Delhi.