Authors:1*Sunanda Biswas and 2Bharat H. Gawade
1Division of Soil Science and Agricultural Chemistry, ICAR-IARI, New Delhi-110012
2Quarantine Division, ICAR-National Bureau of Plant Genetic Resources, New Delhi-110012
*Corresponding author, email: sunandabiswas13@gmail.com
Introduction
The diversity in soils is several orders of magnitude higher than that above ground and is seen as the last frontier for biodiversity on earth (Swift, 1999). Biodiversity is the variability among living organisms and the ecological complexes of which they are part, including diversity within species (genetic diversity), between species, and of ecosystems. Soil biodiversity encompasses all organisms that live in the soil, whether they are single-cell organisms or multi-cell animals or plants, which can be classified using traditional taxonomic techniques or genetically via unique DNA or RNA. An understanding of soil biodiversity perspectives in agricultural context is important and useful to arrive at measures that can act as indicators of soil quality and plant productivity. The immense phenotypic and genetic diversity found in soil bacterial and fungal communities makes it one of the most difficult communities to study. It has been suggested that at least 99% of bacteria observed under a microscope are not cultured by common laboratory techniques (Borneman et al., 1996). To overcome problems associated with non-culturable bacteria and fungi, various methods have been developed to identify and study these microorganisms including numerous DNA and RNA-based methods. The calculation of the indices of soil biodiversity is important to indicate the total make up of a community in soil.
Elements of Biodiversity
Biodiversity can be represented by three interrelated elements: genetic, functional and taxonomic diversity.
Genetic diversity
can identify individual organisms from some unique part of their DNA or RNA and so provides definitive information on soil biodiversity. These approaches have already highlighted the immense scale of soil microbial diversity, which has also been recognised for its economic potential through bio prospecting. In other words, it can also be defined as gene and genotype variation within a species. It consists of the whole set of genetic information contained in the genes of all the animals, plants and microorganisms populating the Earth.
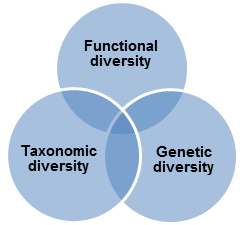
Fig 1: Different elements of biodiversity
Functional diversity reflects the range of ecological services provided by soil organisms and may be expressed in terms of the species or groups important to the maintenance of different soil functions, for example ecosystem engineers that mediate nutrient flows and soil. This approach has focussed on the provision of soil functions for agricultural production or environmental protection but has been modified to address the importance of biodiversity per se in soil resilience and its relevance to nature conservation, heritage interests and other economic uses.
Taxonomic diversity is typically defined by species richness, as simple counts of species recorded or relative differences in species composition. Taxonomic diversity can also be defined at various higher taxonomic levels (e.g. phyla, orders or families). This approach has been proposed as a more realistic level at which, in the first instance, to carry out inter-site comparisons.
Types of diversity among biota
Types of diversity among biota can be broadly classified according to two points of view
- Taxonomic view
- Agricultural microbiological view
- α–diversity: is the diversity of species within a local habitat.
- β–diversity: is the rate of change of diversity along a gradient between habitats and it is also known as regional diversity.
- γ–diversity: is the richness of species within a geographical area (i.e. α+ β diversity)
- Metabolic diversity: Due to major differences between the metabolic needs, heterotrophs v/s autotrophs, and between catabolic styles such as fermentation v/s respiration (aerobic and anaerobic).
- Structural diversity: Due to major structural differences. ex. gram-positive and gram-negative bacteria, (bacteria and archaea), presence or absence of walls, external appendages and endospores etc.
- Morphological diversity: Due to different shapes ex. bacilli, cocci, spirals, filamentous forms, pleiomorphic.
Methods of studying microbial diversity in soil
Methods to measure microbial diversity in soil can be categorized into two groups: biochemical-based techniques (Table 1) and molecular-based techniques (Table 2)
Biochemical-based techniques to study microbial diversity
Most methods described below can be used for either bacteria or fungi, although some are specific to one or the other.
Plate counts
Traditionally, diversity was assessed using selective plating and direct viable counts. These methods are fast, inexpensive and can provide information on the active, heterotrophic component of the population.
Sole carbon source utilization patterns/community level physiological profiling for measuring microbial diversity
In this technique using commercially available 96-well microtitre plate to assess the potential functional diversity of the bacterial population through sole source carbon utilization (SSCU) patterns. Biolog introduced an Eco-plate containing 3 replicates of 31 different environmentally relevant carbon sources and one control well per replicate. CLPP is useful when studying the functional diversity of soils and is a valuable tool especially when used in conjunction with other methods.
Fatty acid methyl ester (FAME) analysis
Fatty acids make up a relatively constant proportion of the cell biomass and signature fatty acids exist that can differentiate major taxonomic groups within a community. Therefore, a change in the fatty acid profile would represent a change in the microbial population. For FAME analysis, fatty acids are extracted directly from soil, methylated and analyzed by gas chromatography. This method will detect changes in the composition of the bacterial and/or fungal community, as well as enable one to follow signature fatty acids of different groups of microorganisms.
Table 1. Advantages and disadvantages of biochemical-based methods to study soil microbial diversity
| Method | Advantages | Disadvantages | Selected references |
| Plate counts | Fast, inexpensive | Unculturable microorganisms not detected | Tabacchioni et al. (2000) |
| CLPP | Fast, highly reproducible | Only represents culturable fraction of community | Classen et al. (2003) |
| FAME | No culturing of microorganisms, direct extraction from soil | If using fungal spores, a lot of material is needed | Zelles. (1999) |
Molecular-based techniques to study microbial Diversity
Guanine plus cytosine (G+C) content
Differences in the guanine plus cytosine (G+C) content of DNA can be used to study the bacterial diversity of soil communities. It is based on the knowledge that microorganisms differ in their G+C content and that taxonomically related groups only differ between 3% and 5% (Tiedje et al., 1999). This method provides a coarse level of resolution as different taxonomic groups may share the same G+C range.
Table 2. Advantages and disadvantages of some molecular-based methods to study soil microbial diversity
| Method | Advantages | Disadvantages | Selected references |
| (G+C) content | Not influenced by PCR biases | Requires large quantities of DNA | Tiedje et al. (1999) |
| DGGE and TGGE | Large number of samples can be analyzed simultaneously, Reliable, reproducible and rapid | PCR biases, dependent on lysing and extraction efficiency, can represent more than one species (co-migration) | Maarit-Niemi et al. (2001) |
| SSCP | Same as DGGE / TGGE, no GC clamp, no gradient | PCR biases, some ssDNA can form more than one stable conformation | Tiedje et al. (1999) |
| ARDRA or RFLP | Detect structural changes in microbial community | PCR biases, banding patterns often too complex | Tiedje et al. (1999) |
| T-RFLP | Compare differences in microbial communities | PCR biases, dependent on lysing and extraction efficiency | Osborn et al. (2000) |
| RISA or ARISA | Highly reproducible community profiles | Requires large quantities of DNA | Fisher and Triplett (1999) |
PCR-based approaches
PCR targeting the 16S rDNA has been used extensively to study prokaryote diversity and allows identification of prokaryotes as well as the prediction of phylogenetic relationships (Pace, 1999). 18S rDNA and internal transcribed spacer (ITS) regions are increasingly used to study fungal communities. In these methods, DNA is extracted from the environmental sample and purified. Target DNA (16S, 18S or ITS) is amplified using universal or specific primers and the resulting products are separated in different ways.
Denaturing gradient gel electrophoresis (DGGE)/temperature gradient gel electrophoresis (TGGE)
Denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) are two similar methods for studying microbial diversity. DNA is extracted from soil samples and amplified using PCR with universal primers targeting part of the 16S or 18S rRNA sequences. On denaturation, DNA melts in domains, which are sequence specific and will migrate differentially through the polyacrylamide gel. Theoretically, DGGE can separate DNA with one base-pair difference. TGGE uses the same principle as DGGE except the gradient is temperature rather than chemical denaturants.
Single strand conformation polymorphism (SSCP)
Another technique that relies on electrophoretic separation based on differences in DNA sequences is single strand conformation polymorphism (SSCP). Single-stranded DNA is separated on a polyacrylamide gel based on differences in mobility caused by their folded secondary structure.
Restriction fragment length polymorphism (RFLP)/amplified ribosomal DNA restriction analysis (ARDRA)
In the study PCR amplified rDNA is digested with a 4-base pair cutting restriction enzyme. Different fragment lengths are detected using agarose or non-denaturing polyacrylamide gel electrophoresis in the case of community analysis. RFLP banding patterns can be used to screen clones or used to measure bacterial community structure.
Terminal restriction fragment length polymorphism (T-RFLP)
Terminal restriction fragment length polymorphism (T-RFLP) is a technique that addresses some of the limitations of RFLP (Tiedje et al., 1999). It follows the same principle as RFLP except that one PCR primer is labelled with a fluorescent dye, such as TET (4,7,2V,7V-tetrachloro-6-carboxyfluorescein) or 6-FAM (phosphoramidite fluorochrome 5-carboxyfluorescein). This allows detection of only the labelled terminal restriction fragment.
Ribosomal intergenic spacer analysis (RISA) and automated ribosomal intergenic spacer analysis (ARISA)
In RISA and ARISA, the intergenic spacer (IGS) region between the 16S and 23S ribosomal subunits is amplified by PCR, denatured and separated on a polyacrlyamide gel under denaturing conditions. In RISA, the sequence polymorphisms are detected using silver stain while in ARISA the forward primer is fluorescently labelled and is automatically detected.
Indices of soil biodiversity
There are some indices of soil biodiversity which are important to indicate the total make up of a community.
Species Richness: It is simply the total number of species.
Species Evenness: It indicates the distribution of the individuals within species designations.
Diversity Indices: It combines species richness and species evenness together, has been called Heterogeneity Indices.
Moreover a diversity index is a single value; thus it cannot indicate the total make up of a community. For example, two communities may have same diversity index value, but one may comprise low evenness and high richness, but other may comprise high evenness and low richness. Therefore, it is necessary to look at all the three indices.
Richness, Evenness and Diversity indices and equations
- Richness
The Margalef and Menhinick equations are based on the assumption that a relationship between S (total no. of species) and n (total no. of individuals) exists. One way to avoid deviating from these assumptions is to use samples of equal size.
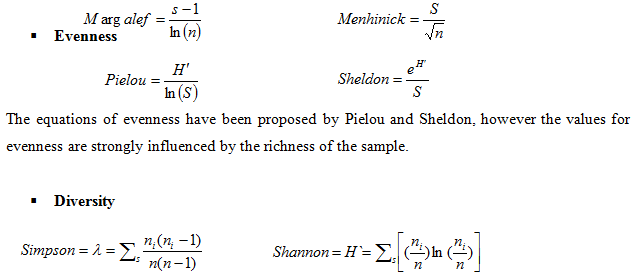
In diversity indices, the Simpson diversity index was the first index to be used in ecological studies, and deals with probabilities that two individuals are of same sp. in other words, if the probability that two individuals are in the same sp. is low, then the diversity is high. The Shannon index deals with the uncertainty predicting to which sp. an individual will belong. This diversity index is most widely used; however it is related to the number of sp. in the sample and does not consider the rare sp. In the equation, ni= importance value of each sp.; N= total of importance value. Shannon Index may also be used to determine functional diversity.
Conclusion
The development of approaches that do not require the establishment of microbial cultures will undoubtedly enhance our knowledge of biodiversity and promote the discovery of new microorganisms with unique capacities for bioremediation, soil restoration, and therapeutic applications. Although much research is still needed, the recognition of the importance of soil biodiversity has taken place and the direction of the developments in protection of biodiversity and its application of soil monitoring and in man-used ecological services is promising.
References:
1. 1. Borneman, J., Skroch, P.W., O’Sullivan, K.M., Palus, J.A., Rumjanek, N.G., Jansen, J.L., Nienhuis, J., and Triplett, E.W.1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol., 62: 1935– 1943.
2. Classen, A.T., Boyle, S.I., Haskins, K.E., Overby, S.T., Hart, S.C. (2003). Community-level physiological profiles of bacteria and fungi: plate type and incubation temperature influences on contrasting sois. FEMS Microbiol. Ecol 44, 319–328.
3. Fisher, M.M., Triplett, E.W. (1999). Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol 65, 4630–4636
4. Margalef, R. (1958). Information Theory in Ecology. Gen. Syst 3, 36-71.
5. Menhinick,E.F. (1964). A Comparison of Some Species Individual Diversity Indices Applied To Samples of Field Insects. Ecology 45, 859-861.
6. Niemi, R. Maarit; Heiskanen, Ilse; Wallenius, Kaisa And Lindström, Kristina. Extraction and purification Of DNA in rhizosphere soil samples for PCR-DGGE analysis of bacterial consortia. Journal Of Microbiological Methods, July 2001, Vol. 45, No. 3, P. 155-165.
7. Osborn, A.M., Moore, E.R.B., Timmis, K.N. (2000). An evaluation of terminal-restriction fragment length polymorphisms (TRFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol 2, 39– 50.
8. Pace, N.R. (1999). Microbial ecology and diversity. ASM News 65, 328– 333.
9. Shannon C. E, and Weaver W. (1949). The Mathematical Theory of Communication. University Illinois Press. Urbana, IL.117p.
10. Simpson, E.H.(1949). Measurement of Diversity. Nature 163,688.
11. Swift, M.J. (1999). Towards the second paradigm: integrated biological management of soil. In: InterrelaçãoFertilidade,Biologia do Solo e Nutrição de Plantas (J.O. Siqueira, F.M.S. Moreira, A.S.Lopes, L.R.G. Guilherme, V. Faquin, A.E. Furtani Neto and J.G. Carvalho, eds.). UFLA, Lavras, Brasil, 11-24.
12. Tiedje, J.M., Asuming-Brempong, S., Nusslein, K., Marsh, T.L., Flynn, S.J. (1999). Opening the black box of soil microbial diversity. Appl. Soil Ecol 13, 109– 122.
Zelles, L. (1999). Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol. Fertil. Soils 29, 111 –129
About Author / Additional Info:
WORKING AS A SCIENTIST IN ICAR-IARI