Authors: Ashok Kumar, Ansheef Ali
Plants represent a rich source of nutrients for many organisms including bacteria, fungi, protists, insects, and vertebrates. Although lacking an immune system comparable to animals, plants have developed a stunning array of structural, chemical, and protein based defenses designed to detect invading organisms and stop them before they are able to cause extensive damage. Humans depend almost exclusively on plants for food, and plants provide many important nonfood products including wood, dyes, textiles, medicines, cosmetics, soaps, rubber, plastics, inks, and industrial chemicals. Understanding how plants defend themselves from pathogens and herbivores is essential in order to protect our food supply and develop highly disease resistant plant species.
Plant diseases or phyto-pathogens (bacteria, fungi, viruses, and pests) affect plants right from the planting stage up to harvesting and storage of the product. For plants to survive the harmful effects of those antagonistic interactions, they must overcome the pathogenic mechanisms that results in the disease process. Consequently, they have evolved several different defense mechanisms by which they defend themselves against these microbial infections. These defense mechanisms or immunity is termed basal resistance, a term referring to two distinct aspects of plant-pathogen interaction: constitutive (physical barriers to pathogens) and induced (non-specific pathogen elicited) defenses.
Plant Immunity
Broadly defined, disease is any physiological abnormality or significant disruption in the “normal” health of a plant. Disease can be caused by living (biotic) agents, including fungi and bacteria, or by environmental (abiotic) factors such as nutrient deficiency, drought, and lack of oxygen, excessive temperature, ultraviolet radiation, or pollution. In order to protect themselves from damage, plants have developed a wide variety of constitutive and inducible defenses. Constitutive (continuous) defenses include many preformed barriers such as cell walls, waxy epidermal cuticles, and bark. These substances not only protect the plant from invasion, they also give the plant strength and rigidity. In addition to preformed barriers, virtually all living plant cells have the ability to detect invading pathogens and respond with inducible defenses including the production of toxic chemicals, pathogen degrading enzymes, and deliberate cell suicide. Plants often wait until pathogens are detected before producing toxic chemicals or defense related proteins because of the high energy costs and nutrient requirements associated with their production and maintenance.
There are two major layers of biochemically induced plant immunity. These comprises; pathogen-associate molecular patterns (PAMPs)-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI is initiated by the perception of microbe conserved PAMPs, such as bacterial flgellin and elongation factor Tu (EF-Tu), with specific plant cell. Surface pattern-recognition receptors (PRRs), PTI usually activates some early resistance responses, including stomatal closure, activation of mitogen-activated protein kinase (MAPK) cascades, transcription of resistance-related genes, reactive oxygen species (ROS) production and callose deposition. ETI on the other hand was thought to a form of accelerated and amplified PTI response. It is activated by plant intracellular resistance (R) genes (proteins) after specific perception of pathogenic effectors, and is associated with programmed cell death, a response which is referred to as the hypersensitive response (HR). Natural selection results in new R specificities so that ETI can be triggered again. So this natural selection will ultimately decide the susceptibility or resistance of host.
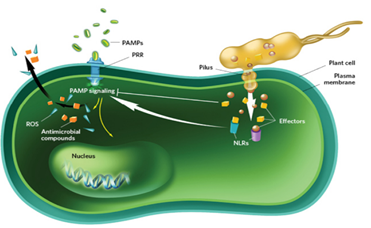
Viruses are well-known to be one of the major concerns for agricultural production and food security throughout the world. It is estimated that viral agents are responsible for the half of emerging diseases reported in plants. The control of plant viruses is often dependent on the use of pesticides; however, such strategy has many adverse environmental effects. In many plant virus-related outbreaks, the disease management is difficult to accomplish due to the variability of factors affecting the development of the disease, such as local climate conditions, plant aging, crop varieties, vector transmission efficiency and severity of viral strains. Unlike other pathogens (i.e., fungi and bacteria), plant viruses cannot be controlled chemically and a combination of cultural practices, biosecurity measures, organism vector management and plant genetic resistance is needed to deal with the disease.
So to generate a plant which is resistant to virus is a difficult task. For that there were basically two approaches, breeding and genetic engineering approaches. Later approach includes number of strategies in which newly arising genome editing tools are highlighting once. These are the tools that can insert, delete, or modify the DNA by using artificially engineered enzymes (site specific nuclease)
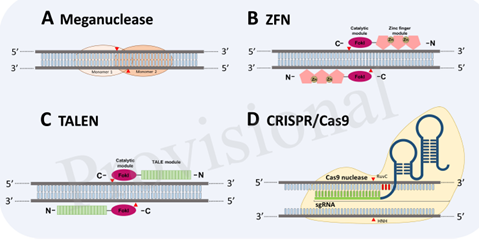
There are four major classes of SSNs: meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced palindromic repeats/CRISPR-associated. These techniques have been harnessed independently to improve crop plant resistance to viruses directly by targeting viral genomes or by targeting host factors. Out these various tools by considering various points like off target cleavage, absence of off targets in plant genome, difficulty in designing construct one can say that CRISPR/Cas9 is the most promising one of these all genome editing system.
CRISPR/Cas9
CRISPR stands for clustered regularly interspaced short palindromic repeats were fortuitously discovered in Escherichia coli in 1987. More than 87% of archaeal genomes and more than 45% of bacterial genomes carry at least one CRISPR locus. These are actually one of the resistance mechanism towards the infecting bacteriophage or extra chromosomal DNA (plasmid). Along with these CRISPR loci, protein encoding CRISPR associated genes are present upstream and reported in almost all loci across the several bacterial genome, which is required for cleavage of target.
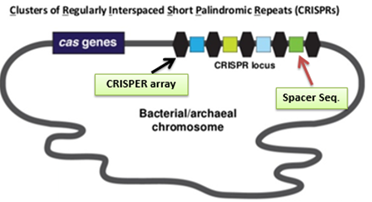
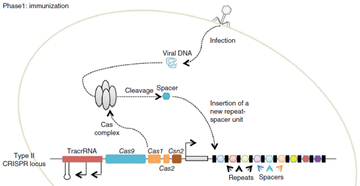
Complete process of this immunity is divided in to two phases
- Immunization In the immunization phase, the CRISPR system stores the molecular signature of a previous infection by integrating fragments of invading phage or plasmid DNA into the CRISPR locus as ‘spacers’. It means whenever there is attack from invading bacteriophage, the cas protein specifically cas1&2 used to target the genome of invader phage, and it save the fragment of DNA(sequence) as spacer at 5’ prime locus of CRISPS array
-
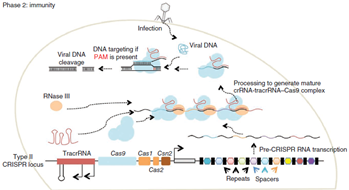
Immunity In the immunity phase, the bacterium uses this stored information to defend against invading pathogens by transcribing the locus and processing the resulting transcript to produce CRISPR RNAs (crRNAs) that guide effector nucleases to locate and cleave nucleic acids complementary to the spacer. First, tracrRNAs hybridize to repeat regions of the pre-crRNA. Second, endogenous RNase III cleaves the hybridized crRNA-tracrRNA, and a second event removes the 5′ end of the spacer, yielding mature crRNAs that remain associated with the tracrRNA and Cas9. The complex cleaves complementary ‘protospacer’ sequences only if a PAM sequence is present. So as mentioned tracrRNA has role in the maturation of crRNA and it stabilize the complete structure and help in activation of cas9 mediated cleavage. Protospacer adjacent motif present downstream of the target site is the pre-requisite for the cleavage by cas9 protein complex. Cas9 protein mainly consist of 2 different domain which is specific for cleavage activity. That are HNH domain that cleaves strand of DNA complementary to gRNA, and Ruvc domain which cleaves non-complementary strand of DNA.
So once a DSB (Double stranded break) produced by CRISPR construct, it can be rejoined or repaired by two basic mechanism. For this HDR (homology directed repair), It is a recombinational repair mechanism of DSB in which gene insertion and gene modification can be achieved. Another mechanism NHEJ (non homologous end joining), it’s an error prone mechanism normally leads to deletion/addition of 1-10bp i.e. indel mutation. Geminiviridae are a family of DNA viruses that infect a diversity of plants. These insect-transmitted viruses can cause destructive diseases in crop plants and have been described as a curse to food security. Until now, limited progress has been made with developing crop varieties resistant to geminiviruses. Ji et al. report on a new strategy towards improving plant resistance to geminiviruses using the bacterial CRISPR/Cas system. In this current study they proved the CRISPR derived immunity in plants towards BSCTV (Beet severe curly top virus) i.e a geminivirus which generally attacks dicot plants like tomato, cassava, cotton, sugar beet pepper etc. in study 2 plant species are used Nicotiana and Arabidopsis and developed transgenic lines of these plants having CRISPR derived immunity towards BSCT virus. For constructing sgRNA targets they selected 3 regions in genome. That are coat protein, origin of replication and replication initiator protein. Each region is off 300bp in length and in each region they randomly selected 13-15 target sites which are having PAM recognition site for cleavage. And constructed a total of 43 sgRNA-CRISPR construct. Agrobacterium harboring this construct containing vector pHSN401 used for agroinoculation during transient analysis. BSCTV containing another vector i.e. pCAMBIA used for viral transfer. There are several advantages of using CRISPR/Cas to confer geminivirus immunity in plants. CRISPR/Cas allows simultaneous targeting of a single or multiple genetic loci in one or several Gemini viruses. The simplicity and robustness of the CRISPR/Cas technology will make it possible to respond to newly emerging strains by deploying appropriate sgRNA transgenes into a crop. However, despite clear advantages, the use of the CRISPR/Cas technology for engineering Gemini virus resistant crops is associated with significant challenges. First, transgenic crops expressing CRISPR/Cas may not be favorably perceived by regulators resulting in high commercialization costs. As a consequence, the strategy of using CRISPR/Cas for resistance to Gemini viruses may be commercially viable for major field crops, such as maize, but not cost-effective for crops grown on a smaller scale, such as tomato. Second, constitutive expression of Cas9 and sgRNA(s) may result in off-target mutations in the crop genome that may build up over time. Also, the strategy of using several sgRNAs to target multiple viruses may further increase the rate of off target mutations, and the guide sequence within the sgRNA transgene may mutate so additional off-target mutations may get introduced. Overall, we need to better understand the rate and degree to which CRISPR/ Cas off-target mutations arise in plants. An intriguing question is whether expressing CRISPR/ Cas in crops will exert an enormous selection pressure on geminiviruses and, as a result, accelerate their evolution. The CRISPR/Cas system may select for synonymous or neutral nonsynonymous mutations in targeted coding sequences that would enable the virus to escape cleavage. It should be pointed out that the CRISPR/Cas system is mutagenic by nature and thus acceleration of virus evolution can be expected. The CRISPR/Cas system may also enhance recombination between distinct Gemini viruses when plants get infected by multiple virus strains. Such recombinogenic effect is another risk factor that needs to be taken into account.
References:
1) Ji, X., Zhang, H., Zhang, Y., Wang, Y., & Gao, C. (2015). Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nature Plants, 1, 15144.
2) JH, D. (2015). An Overview of Plant Immunity. Plant Pathology & Microbiology.
3) Jones, J. D., & Dangl, J. L. (2006). The plant immune system. Nature, 444(7117), 323-329.
4) Doudna, J. A., & Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096.
5) Chaparro-Garcia, A., Kamoun, S., & Nekrasov, V. (2015). Boosting plant immunity with CRISPR/Cas. Genome biology, 16(1), 254.
About Author / Additional Info:
We are currently pursuing Ph.D in Biochemistry from Indian Agricultural Research Institute.