Authors: Vijaykumar I. Prajapati* and Harshvardhan N. Zala
Department of Agricultural Biotechnology
Anand Agricultural University, Anand.
Introduction:
Genetic engineering of plants mostly involves the addition of genetic material (single or multiple genes) that is integrated into a recipient plant, leading to the modification of the plant’s genome. The plants with modified genome are known as transgenic plants or genetically modified (GM) plants. Selectable marker gene systems are vital for the development of transgenic crops. Since the creation of the first transgenic plants in the early 1980s and their subsequent commercialization worldwide over almost an entire decade, antibiotic and herbicide resistance selectable marker gene systems have been an integral feature of plant genetic modification. Without them, creating transgenic crops is not feasible on purely economic and practical terms.
However, after selection of transformed plants, the presence of marker genes in their genomes becomes useless and their effects become unpredictable or even potentially dangerous. So in these contrasts the novel technology development for creation of transgenic plants devoid of “genetic waste”, e.g., selectable markers and reporter genes.
What is Transgenic Plants?
The Plants with modified genome are known as transgenic plants. It usually normal in appearance and character differ from the normal plant with respect to function and influence of the inserted gene.
What is Selectable Markers?
A gene identified cells that have been transformed or transfected with a vector is called as Selectable Markers e.g. antibiotic resistance genes and herbicide resistance genes etc. Presence of a suitable marker is necessary to facilitate the selection of few transformed cells and the detection of transgenic plants during their development. These selectable marker genes produce a phenotype that enables only the plants that contain and express this gene to survive when the selection pressure imposed on them.
The Key Issues related to Selectable Marker Gene
Antibiotic Resistance Genes:
(i) Effects on the therapeutic efficacy of clinically used antibiotics: antibiotic resistance gene products in transgenic crops or products might render clinically important therapeutic antibiotics ineffective.
(ii) Potential for horizontal gene transfer: Concerns about the potential transfer of the antibiotic resistance marker genes to intestinal and soil microorganisms.
Herbicide Resistance Genes:
(i) Gene flow: By which new genes can spread by normal out-crossing to wild or weedy relatives of the engineered crop and become Superweeds.
(ii) Weediness: The potential for a crop or its sexually compatible wild relatives to become established and to persist and spread into new habitats as a result of newly introduced genes.
(iii) Toxicity and Allergenicity: It is associated with human health and the safety of novel foods and potential negative effects on non-target organisms.
Approaches for production of Selectable Marker Gene (SMG) free transgenic plants
• Avoidance of Selectable Marker Gene (SMG)
• Integration of SMG and GOI on unlinked genomic loci-during transformation so that the SMG can be segregated away from the GOI (Gene of Interest)
• Integration of SMG and GOI on the same locus, but molecular tools are used for SMG removal
Strategies to develop Marker-Free Transgenic
1. Positive selection system: Identification and selection of transformed cells without injury or death of the non-transformed cells. The selectable marker genes code for enzymes that give the transformed cells a capacity to metabolize specific nutrient substances not usually metabolized by normal plants. Addition of this substance in the culture medium, as nutrient source during the regeneration process, allows normal to enhanced growth and differentiation of transformed cells while non-transformed cells will not be able to grow and regenerate.
2. Negative selection system: In this method places negative selectable marker genes next to a positive marker gene in the same construct and are a powerful means of creating marker-free transgenic plants. Transformed plants are selected for the absence of negative selection marker genes under the selection pressure of a negative selection Marker. This allows for the selection of marker-free transgenic plants without performing a large number of PCRs.
3. Abiotic stress related genes as selection markers: It is a well-known fact that various genes encode proteins that protect the plants at the time of several environmental stresses like drought stress, salt stress and oxidative stress. Till date so many genes that are well characterized in A. thaliana or in several agronomically important crops can be used for the development of marker-free transgenic plants. The basic ideas behind this strategy is that plant tissue or the plant senses high Na+ concentration in the soil/media and initiates signal transduction to activate a set of stress responsive genes for salt tolerance. The gene is incorporated into the plant tissue or explants without the selection marker gene. After transformation the tissue will grow under the pressure of salt stress, and explants that grow well without any deformities are selected and grown further in the salt stress medium until the vegetative proliferation of the explants. In the whole experiment there is no need to use the selection marker. The gene itself can be used as selection marker to select the transformed tissue
4. Avoiding the use of selectable marker genes: The simplest way to eliminate marker genes from transgenic is to avoid their use in the transformation of plants.
Strategies to eliminate Marker Genes from Transgenic:
• Co-transformation and Segregation
• Transposon based marker methods
• Site-Specific Recombination
• Multi-auto transformation (MAT) vector system
• Auto-excision strategy
• Intra-chromosomal Recombination (ICR) system
Recently developed techniques for removal of SMGs from Transgenic
1. Mega-nuclease based SMGs removal
2. Zinc finger nuclease (ZFN) technology for SMGs removal
3. Transcription activator-like effector (TALE) for SMGs removal
4. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) for SMGs removal
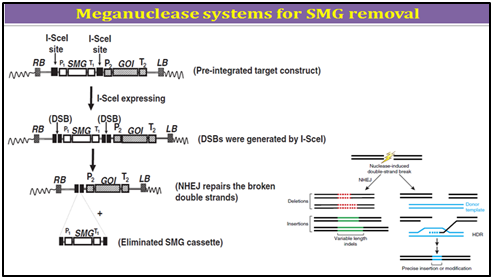
1. Mega-nuclease based SMGs removal
Mega-nucleases are homing endonucleases, a large family of DNA nucleases found in eukaryotes, bacteria and archae-bacteria. Homing endonucleases typically are encoded by introns and inteins. Similar to type II restriction enzymes, they cleave double-strand DNA. However, while type II restriction enzymes recognize short nucleotide sequences (3�"8 bp), homing endonuclease recognize large target sequences (12�"40 bp), which are long enough to occur randomly only with a very low probability. For example, there is approximately one I-SceI cutting site every 7�-1010 bp. The low probability of the presence of endogenous restriction sites decreases the risk of genome fractionation by the mega-nuclease upon expression.
I-SceI is one of the common-used homing endonucleases in research, encoded by the mobile group I intron of the large rRNA gene of Saccharomyces cerevisiae, which recognize and cleave a recognition sequence of 18 bases (TAGGGATAACAGGGTAAT). Early studies of the mega-nuclease I-SceI have illustrated how the cleavage activity of this protein initiates homologous recombination events in living cells and demonstrated the recombinogenic properties of chromosomal double-strand breaks or DSBs and is now often used to induce gene targeting by promoting homologous recombination through creating site-specific cleavage, a DSB. In mammalian and plant cells, this approach has enhanced gene-targeting frequency by several orders of magnitude. In addition to use in gene targeting, mega-nuclease engineering was also reported for transgene deletion. By using two I-SceI cutting sites flanking a SMG in a transgene construct, it is possible to induce double-strand breaks on both restriction sites to release the SMG upon the expression of I-SceI. The cut ends were re-joined through the non-homologous end-jointing (NHEJ) repair pathway, the predominant DSB repair pathway in plants.
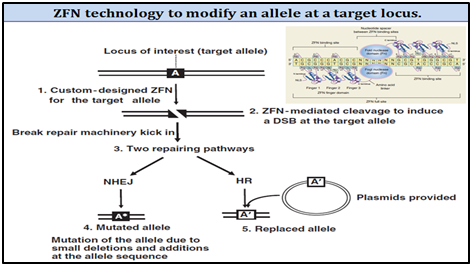
2. Zinc finger nuclease (ZFN) technology for SMGs removal:
ZFNs are referred to as hybrid restriction enzymes. They are fusions of zinc-finger-based DNA recognition modules to endonuclease domains from restriction enzymes. These enzymes are engineered to contain a zinc finger DNA-binding domain (composed of 3�"6 zinc fingers) and the nonsequence-specific DNA-cleavage domain from the FokI type II restriction endonuclease (Fn domain). The cleavage domain of ZFNs must dimerize in order to cut DNA; therefore, efficient cleavage requires two zinc-finger-binding sites be located in close proximity to one another.
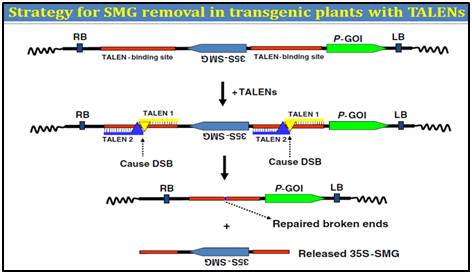
3. Transcription activator-like effector (TALENs)
Transcription activator�"like effector (TALE) proteins are produced by the plant pathogen Xanthomonas. In plants, TALEs localize to the nucleus and bind to target promoter region to trigger the expression of plant genes. Due to their specific DNA binding properties, it fused the restriction domain of FokI nuclease to these TALEs to create TALE-FokI nuclease (TALEN) hybrid proteins. TALENs cause DNA double strand breakage (DSB) at a specific locus. Two identical sets of TALEN-binding sequences can be designed to flank a SMG in a transformation vector.
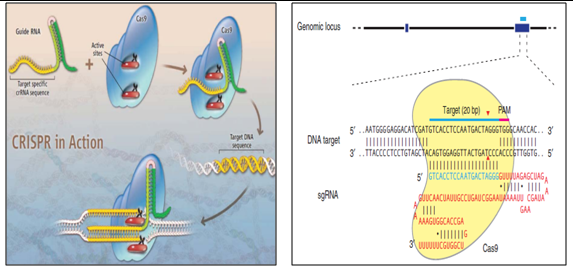
4. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) for SMGs removal:
CRISPR are loci that contain multiple short direct repeats, and provide acquired immunity to bacteria and archaea. Destruction of invading DNA is achieved by an RNA-directed process in which RNAs encoded by the short palindromic repeats of the CRISPR region are transcribed into CRISPR RNA (crRNA) segments that are processed into short, functional RNA fragments with the aid of trans-activating RNA (tracrRNA). Once processing is complete, both crRNA and tracrRNAs become associated with a Cas protein. It contains two nuclease domains. Result in a double-stranded DNA break (DSB) at a site on the foreign DNA just upstream of a protospacer adjacent motif (PAM).
Conclusion:
Various approaches for removal of selectable markers from transgenic have been described here but appropriate choice depends on ease of use, crop and methods of transformation and generation time. The marker free methodologies are useful to overcome the gene flow from the genetic engineered plants to other living organisms. The marker gene should be removed after next generation in order to avoid their toxicity to other living organisms.
References:
1. Ramessar K., Peremarti A., Sonia., Naqvi S. Marian M., Pilar M., Capell T. and
2. Ratner R. (1989) Bio/Technology. 7(4): 337-341.
3. Rukavtsova, Lebedeva A., Zakharchenko N. and Buryanov Ya. (2013) Russian Journal of Plant Physiology 60(1): 14�"26.
4. Tuteja N., Verma S., Sahoo R. Raveendar S. and Reddy (2012) J. Biosci. 37(1): 167�"197.
5. Upadhyaya C., Nookaraju A., Gururani M., Upadhyaya D., Doo-Hwan and Se Won (2010) Botanical Studies 51: 277-292.
About Author / Additional Info:
I am pursing Ph.D. in the Plant Molecular Biology & Biotechnology and also working as Junior Research Fellow at Anand Agricultural University, Anand.