Authors: Lalbahadur Singh, Dr. Nimmy M.S.
Structure of Cas 9
Cas 9 adopts a bilobed architecture composed of a nuclease (NUC) lobe and an α-helical recognition (REC) lobe (Figure 1). The NUC lobe contains the HNH nuclease domain, the RuvC-like nuclease domain, a PAM-interacting (PI) domain, and an evolutionarily divergent wedge domain (WED). The RuvC and HNH nuclease domains use, respectively, a two-metal mechanism and a single-metal mechanism to cleave each of the DNA double strands. The PI domain interacts with the PAM region of DNA through base-specific interaction and contributes to the DNA target specificity of Cas. The WED domain is important for orthogonal recognition of sgRNA scaffolds, and it also interacts with the backbone of the PAM region.
Both biochemical and structural studies have revealed that Sp Cas9 undergoes a series of conformational changes to activate its DNA cleavage activity.
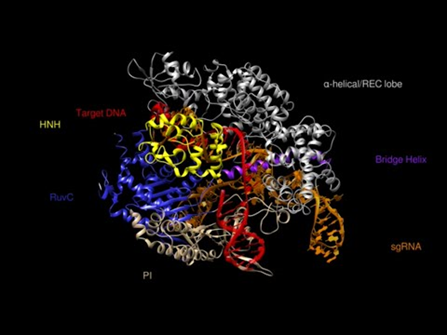
Figure 1. structure of cas 9 protein.
Mechanism of Cas 9 action
A working mechanism of Cas9 has been proposed by combining structural studies and in vitro assays. The Cas9 protein maintains an autoinhibited conformation when not bound by sgRNA in which the active sites in the HNH domain are blocked by the RuvC domain.The binding of an sgRNA induces a conformational change to create a central channel between the two lobes for DNA binding, thus entering into a DNA recognition competent state .The resulting Cas9-sgRNA pre targeting complex can survey DNA for PAMs by three-dimensional diffusion. The Cas9-sgRNA complex binds to a PAM through its PI domain, which initiates local DNA strand separation in the PAM-proximal region to facilitate sgRNA-DNA heteroduplex formation. The Cas9-sgRNA complex will continue to unwind the DNA only if there is a significant match between the guide RNA segment and the target DNA. The strong guide RNA-target DNA base-pairing interactions further promote DNA double strand separation and RNA-DNA heteroduplex formation, which proceeds from the PAM-proximal region and forms a complete R loop. Finally, the complete R loop causes another conformational change in the HNH domain, activating the nuclease activity of both the RuvC and HNH domains to induce DNA cleavage. Sp Cas9 creates a DSB approximately 3 nt away from the PAM in the target DNA. The sgRNA directs Cas9 to a specific genomic locus where Cas9 creates a DSB, which triggers DNA repair through intrinsic cellular mechanisms, such as non homologous end joining or homology-directed repair.
References:
1. Wang, H., Russa, M. L., Qi, L.S. (2016). CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem., 85, 227-64.
2. Jiang, F., Zhou, K., Ma, L., Gressel, S., Doudna, J.A. (2015). A Cas9-guide RNA complex preorganized for target DNA recognition. Science, 348,1477-81.
About Author / Additional Info:
Ph.D. scholar, biotechnology, Indian agricultural research institute (IARI), new delhi. Currently I am working in the area of miRNAs in pulse crop.