Authors: Jyoti Prakash Sahoo1* and Sandeep Kumar Singh2
Ph.D Research Scholar, 1Department of Agricultural Biotechnology, 2Department of Plant Breeding and Genetics, College of Agriculture, OUAT, Bhubaneswar - 751003
*Correspondence mail id : jyotiprakashsahoo2010@gmail.com
RNA silencing
In virus–plant interactions, one of the major mechanisms for plant antiviral immunity relies on RNA silencing. RNA silencing is a nucleotide sequence-specific process that induces mRNA degradation or translation inhibition at the post-transcriptional level (named PTGS in plants) or epigenetic modification at the transcriptional level, depended on RNA-directed DNA methylation (a process named RdDM in plants). RNA interference (RNAi) is a novel phenomenon that has the potential to become an extremely powerful tool for gene silencing in any organism. The process was discovered by Fire et al. (1998). RNA silencing, referred to as gene quelling in fungi and RNA interference (RNAi) in animals. In 1998, Andrew Fire of Carnegie Institute of Washington & Craig Mello of the University of Massachusetts & their colleagues conducted an experiment on the nematode C. elegans. They injected these worms with several different preparations of RNA hoping to stop production of a particular muscle protein. One of the preparations contained “sense” RNA, that is, an RNA having the sequence of the mRNA that encoded the protein being targeted; another preparation contained “antisense” RNA, that is, an RNA having the complementary sequence of the mRNA in question; and a third preparation consisted of a double-stranded RNA containing both the sense and antisense sequences bound to one another. Neither of the single-stranded RNAs had much of an effect, but the double-stranded RNA was very effective in stopping production of the encoded protein. Fire and Mello described the phenomenon as RNA interference (RNAi). They demonstrated that double-stranded RNAs (dsRNAs) were taken up by cells where they induced a response leading to the selective destruction of mRNAs having the same sequence as the added dsRNA.
Different classes of small RNAs:
- MicroRNAs (miRNA): These are endogenous approx. 22 nt RNAs that can play important regulatory roles in animals and miRNAs by targeting mRNAs for cleavage or translational repression. MiRNAs comprise one of the more abundant classes of gene regulatory molecules in multicellular organisms and likely influence the output of many protein-coding genes.
- Small interfering RNAs (siRNA): These are approx. 21-24nt RNA, both endogenous and exogenous origin that can play important antiviral role in both plants and animal. siRNA perform transcriptional nuclear gene silencing through DNA and/or histone methylation.
- Piwi associated small interacting RNAs (piRNA) : These are approx. 30 nucleotides in length and are found in the germline of flies and vertebrates—piRNAs are Dicer independent (Zamore, 2007). PiRNA forms an innate immune system that discriminates transposons from endogenous genes and selectively silences the former. piRNAs enforce silencing by directing transposon DNA methylation during male germ cell development. As such, piRNAs represent perhaps the only currently known sequence-specific factor for deposition of methylcytosine in mammals.
Key Players:
- Dicer (RNase III): Dicer is a special class of RNaseIII family enzyme which specifically binds to double stranded RNA (ds RNA) molecule and make cuts at specific positions to generates small RNA molecules of various length(~20-31nt) type of dicer takes part. Dicing requires both enzyme & Mg2+ to produce siRNAs of 25-27 nt. in length. Dicer contains a dsRNA binding PAZ domain. Glutamte & Aspartate residue perform cleavage in active site of RNase III domain. A distance of 17.5 Å between the RNase III domains fits the width of the major groove of dsRNA. A distance of 65 Å between the PAZ & RNase III domains corresponds to the length of an siRNA fragment containing 25 bp. 3' two nucleotide overhang is recognize by PAZ domain and is conserved across many species.
- Argonaute protein: Argonaute protein family is key players in gene-silencing pathways guided by small RNAs. Small RNAs such as short interfering RNAs (siRNAs), microRNAs (miRNAs) or Piwi-interacting RNAs (piRNAs) are anchored into specific binding pockets and guide Argonaute proteins (AGO) to target mRNA molecules for silencing or destruction. AGO is localized in both cytoplasm & nucleus (in Cajal Bodies). A. Thaliana has 10 Argonaute genes, Ago1 act as a slicer that uses both miRNAs & siRNAs as guides. Ago4 directs siRNA-guided DNA methylation of chromatin.
- RISC Complex: The cytoplasmic RNA-induced silencing complex (RISC) contains dsRNA binding proteins, including protein kinase RNA activator(PACT), transactivation response RNA binding protein (TRBP), & Dicer, that process pre-microRNAs into mature microRNAs (miRNAs) that target specific mRNA species for regulation. The cytoplasmic RISC proteins PACT, TRBP, & Dicer are steroid receptor RNA activator (SRA) binding NR(Nuclear receptor) coregulators that target steroid-responsive promoters & regulate NR activity and downstream gene expression. Size-exclusion chromatography suggested several possible sizes for the RISC, ranging from 500 to 360kD to 140 kD, depending on the model system.
- RNA dependent RNA polymerase (RdRP): RDR helps in synthesis of endogenous long dsRNA precursor of siRNA using ssRNA as template. In Arabidopsis, RDR6 play these role among 6 different RDR proteins. In plants, siRNA of 24nt (DCL3) processed from dsRNA synthesized by RDR2 which plays important role in RdDM pathway.
General pathway:
Of special importance to animals and plants is a type of short noncoding RNA called microRNA (miRNA). miRNAs base-pair with specific mRNAs and regulate their stability and their translation. The miRNA precursors are synthesized by RNA polymerase II and are capped and polyadenylated. They then undergo a special type of processing, after which the miRNA is assembled with a set of proteins to form an RNA-induced silencing complex or RISC. Once formed, the RISC seeks out its target mRNAs by searching for complementary nucleotide sequences. This search is greatly facilitated by the Argonaute protein, a component of RISC, which displays the 5' region of the miRNA so that it is optimally positioned for base-pairing to another RNA molecule. In animals, the extent of base-pairing is typically seven nucleotide pairs, and it usually takes place in the 3'UTR of the target mRNA. Once an mRNA has been bound by an miRNA, several outcomes are possible. If the base-pairing is extensive, the mRNA is cleared by the Argonaute protein, effectively removing its poly-A tail and exposing it to exonucleases. If the base-pairing between the miRNA and the mRNA is less extensive, Argonaute does not slice the mRNA; rather, translation of the mRNA is repressed and the mRNA is destabilized. This effect is associated with shortening of the poly-A tail and the movement of the mRNA to cytosolic structures called processing bodies (P-bodies). Here the mRNAs are sequestered from ribosomes and eventually decapped and degraded.
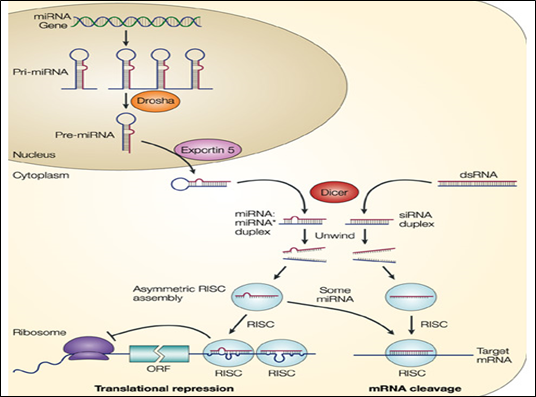
Figure 1: Pathway of miRNA in RNA silencing
RISC Assembly: siRNAs and miRNAs are born double-stranded: the RNase III enzymes, Drosha and Dicer, process the precursor RNAs into small RNA duplexes called siRNA duplexes and miRNA–miRNA* duplexes. Because small RNAs in RISC must anneal to their target RNAs, the two strands of a small RNA duplex must be separated and one strand discarded to form a functional RISC. The discarded strand is called the ‘passenger strand’ whereas the strand retained in RISC is called the ‘guide strand’. RISC assembly can be divided into at least two successive steps: (i) RISC-loading, in which small RNA duplexes are inserted into Ago proteins; and (ii) strand dissociation or unwinding, in which the two strands are separated within the Ago protein. An Ago complex bound to an RNA duplex is called a pre-RISC, whereas a complex containing a single stranded guide is called a mature RISC, holo-RISC or simply RISC. Selection of a single strand from a small RNA duplex is not random; instead, one of the two strands is preferred to serve as the guide. Such asymmetric strand selection depends on the relative thermodynamic stability of the first 1–4 bases at each end of the small RNA duplex; the strand whose 50 end is less stable serves as the guide whereas the other strand is discarded.
RNA silencing as an antiviral defense mechanism in plants:
The majority of plant viruses are positive-sense RNA viruses, which have either ss- or dsRNA genomes and depend on viral RNA dependent RNA polymerase for multiplication. Infection of plants with viruses leads to accumulation of viral siRNAs & in turn, can direct silencing against the viral genome (Ruiz-Ferrer and Voinnet 2009; Voinnet 2005). Host encoded RDR are required for producing the bulk of viral siRNAs from some RNA viruses. 3 RDR in Arabidopsis have been functionally characterized. RDR6 is involved in the tasiRNA pathway, and RDR2 is required for the production of 24-nt siRNAs associated with RNA directed DNA methylation (RdDM). RDR1 appears to play a dominant role in antiviral silencing with RDR6 being also required (Qu 2010). Infection with RNA viruses synthesized 21nt & 22nt siRNA and processed by DCL4 & DCL2, respectively. DCL4 plays a more dominant role than DCL2 in antiviral defense. These 21- and 22-nt viral siRNAs binds to AGO proteins and direct silencing of viral RNAs. AGO1 is the dominant AGO in viral siRNA-directed antiviral defense. In DNA viruses (germini viruses), 21- and 22-nt siRNAs correspond to the coding regions of the viral genome capable of directing RNA cleavage. 24-nt siRNAs are derived from intergenic regions that can direct Cm in this viral genome. The RdDM pathway is involved in plant resistance to DNA viruses.
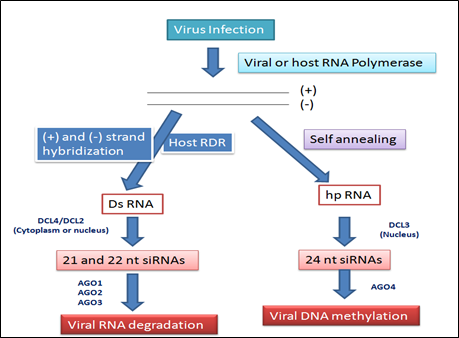
Figure 2: Possible viral small interfering (si) RNA biogenesis pathways
Viral Counter Defense:
Virus has evolved to encode suppressors of RNAi (VSRs), which disrupt RNAi at various steps. VSRs efficiently inhibit host antiviral responses by interacting with the key components of cellular silencing machinery, often mimicking their normal cellular functions. The fact that most viruses have evolved VSRs underlines the antiviral nature of RNA silencing and reveals a pathogen counter defensive strategy with the active suppression of host surveillance. The various VSRs are able to target all effectors of the silencing pathway, such as viral RNA recognition, dicing, RISC assembly, RNA targeting and amplification. Most identified VSRs are multifunctional: besides being RNA-silencing suppressors, they often perform essential roles by functioning as coat proteins, replicases, movement. Proteins, helper components for viral transmission, proteases or transcriptional regulators. This multifunctional nature of VSRs often causes serious difficulties in the exploration of their mechanisms because the inactivation of VSRs often leads to a loss of viability of the given virus.
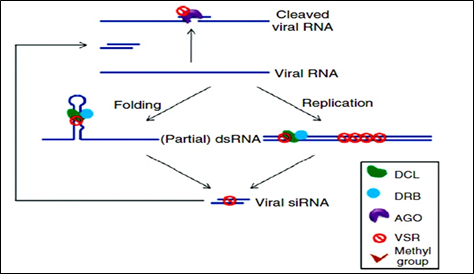
Figure 3: Mechanism of VSRs action.
References:
1. Ding, S.W., Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130: 413-426
2. Ding, S.W. (2010). RNA-based antiviral immunity. Nature Review Immunology 10: 632-644
3. Zorzatto, C., Machado, J.P.B., Lopez, K.V.G. et al., (2015). NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 520: 679-682
About Author / Additional Info: