Authors: BY AASTHA ACHARYA
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) / Cas (CRISPR-associated) system found in most bacteria and Achaea is the RNA mediated adaptive immune that guards the organisms against the viral DNA. Cas 9 is the part of the CRISPR system that grubs, scissors and disrupts the viral DNA in a specific way. The fuction of Cas 9 can be executed via the genetic engineering technique to delete or insert the specific DNA fragment which can be used to change the DNA sequence in human embryo and even remove the integrated human viral HIV DNA. This technique can be used not only in adult cells but also in embryo.
Figure : Storrs; A CRISPR Fore-Cas-t; (2014) The Scientist magazine; http://www.thescientist.com/?articles.view/articleNo/39239/title/A-CRISPR-Fore-Cas-t/
INTRODUCTION
History
In a parallel but completely separate area of research, a few microbiology and bioinformatics laboratories in the mid-2000s began investigating CRISPRs (clustered regularly interspaced palindromic repeats), which had been described in 1987 by Japanese researchers as a series of short direct repeats interspaced with short sequences in the genome of Escherichia coli . The concrete perception regarding the origin of the CRISPRs from plasmid and viral origin was established 2005. The first experimental evidence of the CRISPR-Cas mediated adaptive immune system was shown in 2007 which included the infection experiments of the lactic acid bacterium Streptococcus thermophilus with the involvement of lytic phages. In 2008, mature CRISPR RNAs (crRNAs) were shown to serve as guides in a complex with Cas proteins to interfere with virus proliferation in E. coli. The same year, the DNA targeting activity of the CRISPR-Cas system was reported in the pathogen Staphylococcus epidermidis. Following the 2012 publication of Jinek et al, three studies in January 2013 demonstrated that CRISPR-Cas9 represents an efficient tool to edit the genomes of human cells.
Introduction
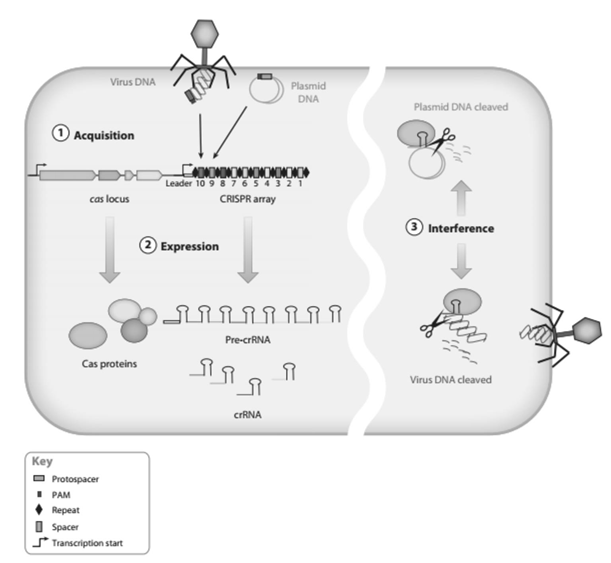
The CRISPR–Cas9 system provides a flexible and modular platform for recruiting different functional effector domains to essentially anywhere in the genome .This is mostly achieved by fusingdCas9 to different effector domains. Sometimes, recruitment of multiple effector domains is required for optimal genome-regulatory effectiveness. In CRISPR/ Cas- mediated immunity, bacteria and Achaea react to viral or plasmid attack in the adaptive phase by first integrating short fragments of foreign nucleic acid (proto spacers) into the host chromosome at the proximal end of the CRISPR array.
It provides acquired resistance against viruses and plasmids via uptake of short fragments of invasive DNA. Cas genes typically are present in the vicinity of CRISPR arrays. CRISPR repeat spacer arrays are transcribed into long primary transcripts that are processed further into a set of short CRISPR RNAs (crRNAs) containing a conserved repeat fragment and a variable spacer sequence (guide) complementary to the invading nucleic acid. crRNAs combine with Cas proteins to form an effector complex which recognizes the target sequence in the invasive nucleic acid by base pairing to the complementary strand and induces sequence-specific cleavage, thereby preventing proliferation and propagation of foreign genetic elements. CRISPR/ Cas systems have been categorized into three main types, based on core elements content and sequences. The structural organization and function of nucleoprotein complexes involved in crRNA-mediated silencing of foreign nucleic acids differ among distinct CRISPR/Cas types.
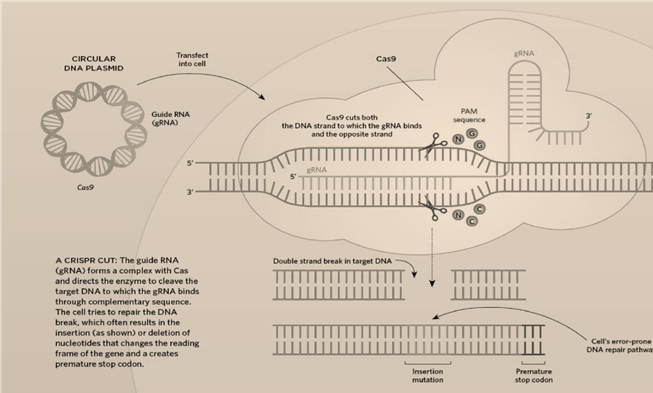
Functionality of CRISPR-Cas 9
Cas9–crRNA complex functions as an RNA-guided endonuclease with RNA-directed target sequence recognition and protein- mediated DNA cleavage. Cas9–crRNA complex functions as an RNA- guided endonuclease that uses RNA for target site recognition and Cas9 for DNA cleavage. Cas9 is functional as a monomer and uses two active sites for the cleavage of opposite DNA strands. Genetic studies showed that S. thermophilus Cas9 is essential for defense against viral invasion, might be responsible for introducing double stranded breaks into invading plasmids and phages, enables in vivo targeting of temperate phages and plasmids in bacteria. In 2012, the S. pyogenes CRISPR-Cas9 protein was shown to be a dual-RNA–guided DNA endonuclease that uses the trans inactivating crRNA: crRNA duplex to direct DNA cleavage. Cas9 uses its HNH nuclease domain (carboxy terminus) to cleave the DNA strand that is complementary to the 20nucleotide sequence of the crRNA; the RuvC-like domain (amino terminus) of Cas9 cleaves the DNA strand opposite the complementary strand. The CRISPR- Cas9 system requires only a change in the guide RNA sequence. For this reason, the CRISPR-Cas9 technology using the S. pyogenes system has been rapidly and widely adopted by the scientific com- munity to target,edit,or modify the genomes of a vast array of cells and organisms. The reported collection of Cas9 orthologs constitutes a large source of CRISPR-Cas9 systems for multiplex gene targeting, and several orthologous CRISPR-Cas9 systems have already been applied successfully for genome editing in human cells.
Glimpse on Working Mechanism of CRISPR-Cas 9
On a repeat encounter with a virus, a bacterium can produce a stretch of RNA that matches the viral sequence, using the material in its spacer archive. This ‘guide RNA’ teams up with DNAcutting Cas enzymes, encoded by nearby CRISPR- associated genes, to seek out and ‘cleave’ the matching viral sequences, stopping the virus from replicating. By engineering the guide RNA, researchers can programme Cas enzymes — most commonly Cas9 — to match the DNA at specific sites that they want to cut in a cell’s genome. This triggers a DNA repair that can result in precise sequence changes to the gene of interest.
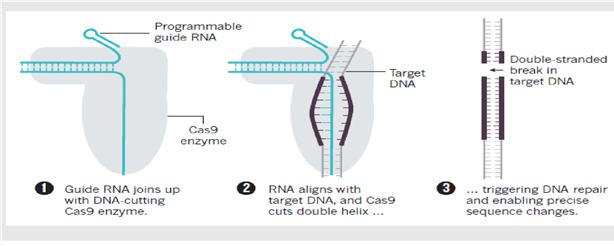
Fig: How CRISPR-Cas 9 Works (24/ 31 December 2015 Vol 528 Nature)
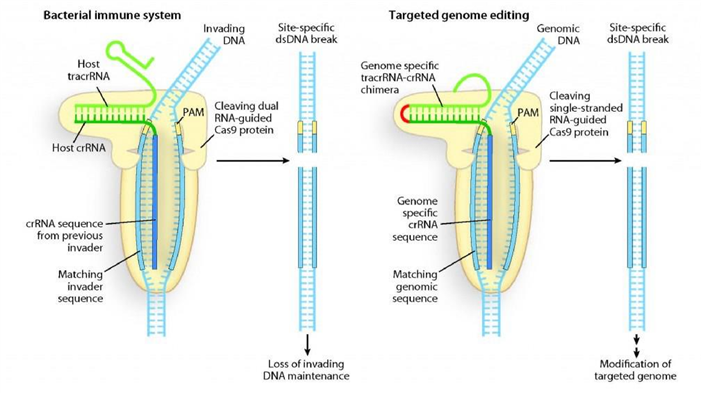
CRISPR-Cas 9 on Genome Editing
Source : lbl.gov; Programmable DNA scissors: A double-RNA structure in the bacterial immune system has been discovered that directs Cas9 protein to cleave and destroy invading DNA at specific nucleotide sequences.
Recently, the type II bacterial CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated) system has been developed as an efficient and versatile technology for genome editing in eukaryotic cells and whole organisms. The hallmark of the CRISPR/Cas system consists of CRISPR arrays composed of spacers interspersed with direct repeats and Cas genes present in the operons. In the expression phase, CRISPR loci are transcribed into precursor CRISPR RNA (pre-crRNA). In the final interference phase, crRNAs are packaged with transactivating crRNA (tracrRNA) and Cas proteins to form ribonucleoprotein complexes that together detect and destroy foreign sequences. The RNA-guided CRISPR-associated protein Cas9 is used for genome editing, transcriptional modulation, and live-cell imaging. Cas9-guide RNA complexes recognize and cleave double-stranded DNA sequences on the basis of 20nucleotide RNA-DNA complementarity, but the mechanism of target searching in mammalian cells is unknown.
The structures of the Cas9–RNA–dsDNA complex show how Cas9 holds unwound double- stranded DNA to enable RNA–DNA hybrid formation without requiring an ATP-dependent helicase activity. DNA binding at a sequence complementary to the RNA segment in the Cas9– RNA complex induces protein structural rearrangements that accommodate both the RNA–DNA helix and the displace non target DNA strand. Those protein nucleic acid interactions in turn direct the non-target DNA strand into the RuvC domain active site, favoring local conformational changes that position the HNH domain active site near the scissile phosphate of the target DNA strand. Cas9 interacts with both ends of the open DNA helix, conferring a 30° helical bend angle that provides the structural distortion required for R-loop stabilization. Collectively, Cas9, an individual nonpolymerase enzyme able to catalyze R-loop formation, achieves this structural change leading to accurate, precise, and programmable DNA cleavage which in turn aids in the editing of the genome.
REFERENCES
- Feng Z, Botao Z, Wona D and Xiaodong L. 2013. "Efficient genome editing in plants using a CRISPR/Cas system." Cell Research 23 p. 1229–1232
- Doudna J. Genome-editing revolution: My whirlwind year with CRISPR. 2015. Nature News. Nature Publishing Group.
- Yang L, Yang LJ, Byrne S, Pan J and Church MG. 2014. CRISPR/Cas9-Directed Genome Editing of Cultured Cells. Current Protocol Molecular Biology. John Wiley & Sons, Inc.
- Gasiunasa G, Barrangoub R, Horvathc P, and Siksnysa V. 2012. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. PNAS. doi:10.1073/pnas.1208507109
- Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, Beheiry ME, Masson JB, Dahan M, Liu Z, Doudna JA, Tjian R. 2015. Dynamics ofCRISPR-Cas9 genome interrogation in living cells. Sciencemag. Vol 350. Issue 626. P823-825
- Jiang F, Taylor DW, Chen JS, Kornfeld EJ, Zhou K, Thompson AJ, Nogales E, Doudna JA.2016. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage.
- Doudna JA, Charpentier E. 2014. The new frontier of genome engineering with CRISPRCas9. Science 346, 1258096 (2014). doi: 10.1126/science.1258096
- Doudna JA and Gerbasch AC. 2015. Genome editing: the end of the beginning. Genome Biology. DOI 10.1186/s13059-015-0860-5
- Du D and Qi LS. 2016. An Introduction to CRISPR Technology for Genome Activation and Repression in Mammalian Cells. Cold Spring Harbor Protocols. doi:10.1101/pdb.top086835
About Author / Additional Info:
I am currently an undergraduate microbiology student from Kathmandu.