Authors: Anirudha Kumar Sahu1 and Prasenjit Debnath2
1,2 M.Sc.(Agri.) in Molecular Biology & Biotechnology,
Dept. of Biotechnology, UAS, Dharwad, Karnataka, PIN-580005
Correspondence mail id: sahuak@uasd.in
Introduction
We all might have heard about the lac operon and typtophan operon system of gene regulation in prokaryotes. Here is another mode of prokaryotic gene regulation system called Riboswitches those come under RNA mediated gene regulation system typically found in bacteria. Recent studies have shown that they are also found in higher plants and certain fungi and also in some archaea. New found classes of riboswitches are being reported at a rate of about three per year.
Basics
Well technically Riboswitches are conserved RNA sequences encoded within the mRNA and are cis-acting regulatory sequences that bind to small entities like metabolites and metal ions as ligands and regulate mRNA transcription or initiation of translation by forming alternative structures in response to the ligand binding. The groups of ligands those can be sensed by Riboswitches include:
1. Metal ions
2. Nucleic acid precursors
3. Enzyme co-factors
4. Amino acids
Mechanism
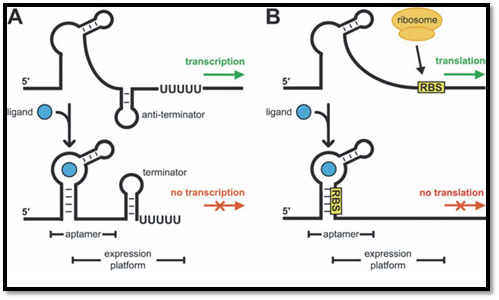
Figure.1) Shows the two major mechanisms of Riboswitch mediated gene regulation in prokaryotes; where A is showing pre-termination of transcription and B is showing blockage of translation initiation by riboswitches.
The Riboswitches are amazing pieces of natural artwork those can regulate a gene at the RNA level by being in the mRNA themselves without the involvement of any other major protein complexes like activators and repressors those are the regular machineries for such regulatory activities. Nearly all examples of the known riboswitches reside in the non-coding regions of the mRNAs. A riboswitch is most commonly located in the 5’ UTR region of a bacterial mRNA. However there are certain exceptions where the riboswitch regulates from a 3’ UTR. Coming to the structure of the Riboswitches, they are basically composed of two domains:
1. The aptamer domain - The Aptamer domain acts as a receptor that specifically binds to a ligand.
2. The expression Platform - The expression platform acts directly on gene expression in response to ligand binding.
There are two ways in which the regulation can happen:
In both the mechanisms the binding of the ligand leads to formation of a hairpin which can lead to structural changes in the nearby residing hairpin and formation of a new hairpin or can take up an important protein binding sequence on the mRNA for the formation of the hairpin which then makes the binding sequence unavailable for protein binding.
1. Pre-termination of transcription - The principle is very simple; binding of the specific ligand to the Aptamer region forms a hairpin like structure that pulls open the anti-termination loop and helps in formation of the termination loop. The formation of the termination loop destabilizes the ternary complex and the RNA Polymerase II dislodges from the template.
2. Blocking translation initiation - This is similar to the above mechanism but the only difference is; this time the binding of the ligand which leads to a hairpin formation takes up the RBS (Ribosome Binding Site) along with it to form the hairpin which makes it unavailable for binding to the Ribosome for the initiation of translation. Thus the translation is blocked. No translation occurs.
Note : There is also a third mechanism of regulation which is not mentioned everywhere in the literatures in which binding of the ligand to the Aptamer leads to formation of the hairpin followed by cleavage at the point of hairpin loop formation and thus rendering the mRNA unavailable for translation.
Riboswitches can also alternate structures that affect the splicing of the pre-mRNA. Sighting one example, a TPP riboswitch (a class of riboswitch) in Neurospora crassa (a fungus) controls alternative splicing to conditionally produce an Upstream Open Reading Frame (uORF), thereby affecting the expression of downstream genes. All these kinds of regulations mentioned above very heavily depend upon the concentration of the ligands present in the surrounding.
Nomenclature of riboswitches
The riboswitches are always named based on the ligand they bind to. Some of the very popular riboswitches are given below those are already functionally validated:
1. Ligand: Metal ions
a) Manganese riboswitches bind manganese ions.
b) Ni Co riboswitches bind the metal ions nickel and cobalt.
c) Magnesium riboswitches bind Magnesium ions
d) Fluoride riboswitches sense fluoride ions, and function in surviving high levels of fluoride.
2. Ligand: Nucleic acid precursors
a) Cyclic di-AMP riboswitches (also called ydaO/yuaA) bind the signaling molecule cyclic di-AMP.
b) Cyclic di-GMP riboswitches bind the signaling molecule cyclic di-GMP in order to regulate a variety of genes controlled by this second messenger
c) Purine riboswitches binds purines to regulate purine metabolism and transport. Different forms of the purine riboswitch bind guanine (a form originally known as the G-box) or adenine.
d) SAH riboswitches bind S-adenosylhomocysteine to regulate genes involved in recycling this metabolite that is produced when S-adenosylmethionine is used in methylation reactions.
e) SAM riboswitches bind S-adenosyl methionine (SAM) to regulate methionine and SAM biosynthesis and transport.
f) TPP riboswitches (also THI-box) binds thiamin pyrophosphate (TPP) to regulate thiamin biosynthesis and transport, as well as transport of similar metabolites. It is the only riboswitch found so far in eukaryotes.
3. Ligand: enzyme co-factors
a) Cobalamin riboswitch (also B12-element), which binds either adenosylcobalamin (the coenzyme form of vitamin B12) or aquocobalamin to regulate cobalamin biosynthesis and transport of cobalamin and similar metabolites, and other genes.
b) FMN riboswitch (also RFN-element) binds flavin mononucleotide (FMN) to regulate riboflavin biosynthesis and transport.
4. Ligand: Amino acids
a) Lysine riboswitch (also L-box) binds lysine to regulate lysine biosynthesis, catabolism and transport.
b) Glutamine riboswitches bind glutamine to regulate genes involved in glutamine and nitrogen metabolism, as well as short peptides of unknown function.
c) Glycine riboswitch binds glycine to regulate glycine metabolism genes, including the use of glycine as an energy source.
d) glmS riboswitch, which is a ribozyme that cleaves itself when there is a sufficient concentration of glucosamine-6-phosphate.
Applications of riboswitches in research and development
1. Development of antibiotics targeting riboswitches.
2. Designing of Ligand-inducible expression systems for common laboratory organisms such as E. coli and B. subtilis which will be more cost effective and simpler than IPTG induced expression system.
3. The ability of a synthetic riboswitch to permit reversible and tunable ligand-dependent gene expression of a protein over its native expression style suggests that synthetic riboswitches may find broad use in studying microbial genetics.
4. Development of more complicated RNA transcription regulation system with more than one riboswitches to develop more control over multiple expression systems; by using more than one ligands (synthetic logic gates) singly or at a time to reprogram cell behavior and to engineer metabolic pathways.
References:
1. Kumar, P. and Mina, U.,2013,Genetics. Life Sciences:Fundamentals and Practice. 2:165pp
2. Topp, S. and Gallivan, J. P., 2010, Emerging Applications of Riboswitches in Chemical Biology. ACS Chem Biol.5(1):139-148.
3. Kim, J. N., Breaker, R. R.,2008, Purine Sensing by Riboswitches. Biology of the Cell. 100(1):1-11.
4. Edwards, A. L., and Batey, R. T., 2010, Riboswitches: A Common RNA Regulatory Element. Nature Education 3(9):9.
5. Garst, A. D., Edwards, A. L. and Batey, R. T., 2011, Riboswitches: Structures and Mechanisms. Cold Spring Harb Perspect Biol. 3(6): a003533.
6. Breaker, R. R., 2012, Riboswitches and the RNA World. Cold Spring Harb Perspect Biol. 4(2): a003566.
7. Macmanus, M., T., non-dated, Riboswitch. Macmanus Lab- illuminating the dark matter of our genome. http://mcmanuslab.ucsf.edu/node/265
8. Anonymous, non-dated, Riboswitches. Boundless.com. https://www.boundless.com/microbiology/textbooks/boundless-microbiology-textbook/microbial-genetics-7/rna-based-regulation-89/riboswitches-483-5373/
9. Anonymous, 2017, Riboswitch. Wikipedia: The free encyclopedia. https://en.wikipedia.org/wiki/Riboswitch
About Author / Additional Info:
Student: M.Sc.(Agri) in Molecular Biology & Biotechnology, IABT, UAS, Dharwad
Research Area: Plant Transformation and screening