Authors: Anshul Watts1, Archana Watts2, Kishor U Tribhuvan1 , Era Vaidya Malhotra1 and Rajendra Prasad Meena1
1 ICAR-National Research Centre On Plant Biotechnology, New Delhi-110012
2 ICAR-Division of Plant Physiology, Indian Agriculture Research Institute, New Delhi-110012
Introduction
Understanding the regulation of gene is one of the most important challenges for molecular biologists. Gene regulation occurs at different levels viz chromatin, transcriptional, post-transcriptional, translational, post-translational etc. Among these transcriptional is the most important level of gene regulation. RNA polymerase binds to specific DNA sequences known as promoter and initiate transcription from a specific site. For some genes transcription starts from multiple sites. Transcription start site (TSS) is the first nucleotide of DNA from where transcription of gene starts i.e. RNA synthesis and it is indicated by +1 sign. In terms of mRNA it is the first nucleotide to which 5ʹ cap (7 methyl guanosine) is attached for stabilizing newly synthesized mRNA. The sequences after the transcription start site in DNA are known as downstream sequences and usually indicated by the + sign. The sequences before the transcription start site are known as upstream sequences and usually indicated by â€" sign.
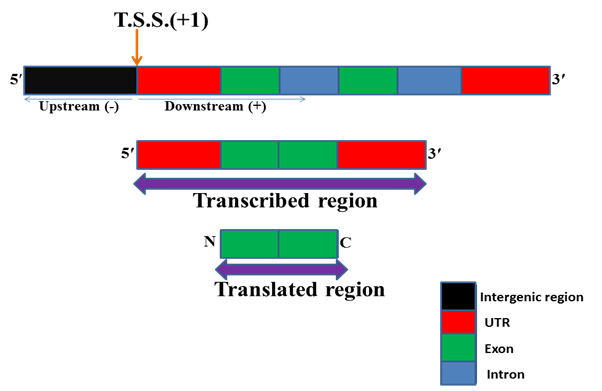
Fig. 1 Structure of a typical eukaryotic gene
Importance and features of TSS
For molecular biology experiments it is important to determine the TSS because it will help in certain aspects like 1) mapping of 5ʹ end of the gene 2) Determining gene structure 3) predicting and locating the promoter of the gene 4) understand role of promoter in gene regulation because RNA polymerase bind upstream to TSS and initiate transcription.
In case of mammalian genome TSS region is well characterized while it is not in the case of plants. However GC skewed (=(C-G)/(C+G)) around the TSS is conserved both in case of Arabidopsis and rice. It is also reported that highly expressed genes are having more GC skew as compared to the genes which are expressing very low. So GC skewed is related to transcription (Fujimori, S. et. al. 2005). Additionally, CpG island which is a important feature of predicting mammalian TSS is not located in the promoter region of the plant genome. When nucleotide features around upstream and downstream TSS were compared from single loci then they were showing differences.
Methods for determination of TSS
1. Primer extension method
In this method total RNA is isolated from the tissue and stage in which gene of interest is expressing then a primer is designed complementary to the RNA and with the help of reverse transcriptase cDNA is synthesized. This ss cDNA is then run on denaturing PAGE gel. The length of the ss cDNA reflects the distance of the TSS from the 5ʹ end of the primer.
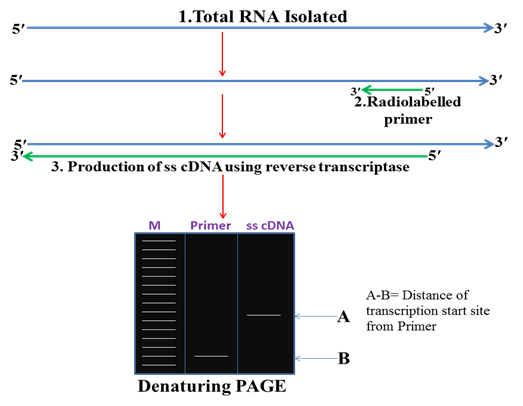
Fig. 2 Primer extension method
2. S1 nuclease mapping method
Mapping of TSS by S1 nuclease is another method used for determining TSS. S1 nuclease cuts single stranded DNA or single stranded RNA but it does not cut double stranded DNA, double stranded RNA or DNA-RNA hybrid. Additionally, this method requires certain information like gene structure, gene sequence, restriction map of the gene and upstream sequence of the gene. Firstly primers are designed from the first exon and several bases upstream to the first exon which is in intergenic region. Using PCR these primers are amplified and then by the help of different restriction enzymes these primers are cut at both ends and radiolabelled at one end. One of strand is degraded while other radiolabelled strand is retained. Then total RNA is isolated from the cell or tissue in which gene of interest is expressing. The radiolabelled DNA probe is hybridized with mRNA and then treated with S1 nuclease. S1 nuclease cuts ssDNA and RNA but not the DNA-RNA hybrids. RNA is degraded in the following step and ss cDNA along with probe is run on a gel. By the comparison of these two bands TSS is then determined.
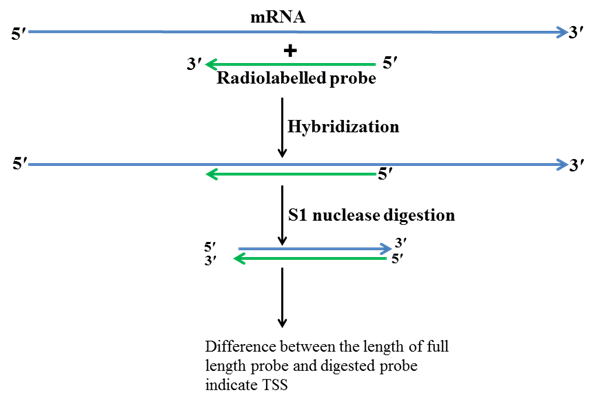
Fig.3 S1 nuclease mapping method
3. 5ʹ RACE (RAPID AMPLIFICATION OF cDNA ENDS)
It is the most common strategy used to determine the TSS. RACE technique was first described by Michael Frohman and Gail Martin. In this technique, first RNA is isolated from the tissue and stage in which the gene of interest is expressing. By use of gene specific primers with the help of reverse transcriptase gene of interest is isolated. Using RNaseA RNA is degraded from the mixture and cDNA is purified. Then with the use terminal deoxynucleotidal transferase enzyme (TdT) and dCTP poly C tail is added at the 3ʹ end of the cDNA. Now this cDNA contains poly C tail at the 3ʹ end. Then an anchor primer is designed, such that it contains poly G and some other arbitrary sequences and some restriction enzyme site for cloning. One primer is designed from gene sequence and the anchored primer is used as second primer for PCR amplification. Another round of PCR can be used using another gene specific primer and anchored arbitrary primer. Then this product is cloned in cloning vector and sequenced. First nucleotide after the G indicates the 5ʹ end of the gene.
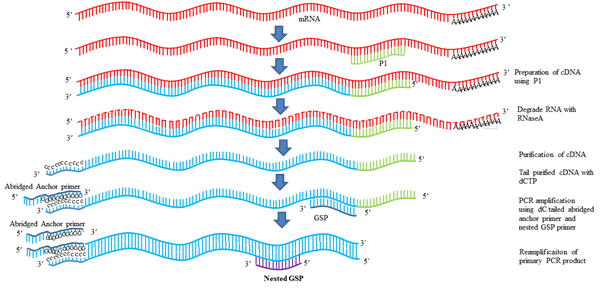
Fig.4 5ʹ RACE for determination of TSS
One of the modification of RACE is RLM-RACE (RNA Ligase Mediated RACE). In this method first RNA is isolated from the tissue and stage in which gene of interest is expressing. Further, this RNA is treated with alkaline phosphatase (CIP) which removes phosphate from 5ʹ end of DNA, rRNA, tRNA, truncated mRNAs. 5ʹ phosphate of all mRNAs is not removed as mRNA contain 5ʹ cap which protects 5ʹ end. Further, this product is treated with Tobacco Acid Pyrophosphatase (TAP) which removes 5ʹ cap from all mRNAs. Then an adaptor containing 20-30 nt known RNA sequence is added in the reaction mixture and with the help of RNA ligase, these adaptor binds to 5ʹ end of the mRNA. Then one primer designed from adaptor sequence and other primer designed from gene is used to amplify the product. This product is further cloned and sequenced. The first nucleotide after the adaptor sequence denotes the TSS.
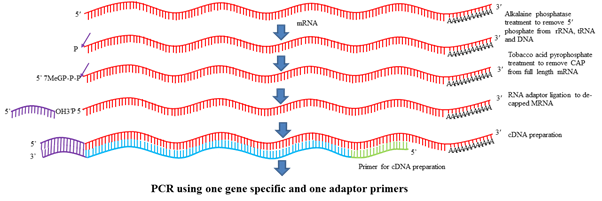
Fig.5 RLM-RACE for determination of TSS
References:
Fujimori S, Washio T, Tomita M. 2005. GC-compositional strand bias around transcription start sites in plants and fungi. BMC Genomics. 6:26
About Author / Additional Info:
I am currently a scientist at National Research Centre On plant Biotechnology, Pusa Campus, New Delhi-110012