Authors: Pooja Kumari**, S.P. Singh* and K. K. Gangopadhyay*
**Scientist, *Principal Scientist,
ICAR-National Bureau of Plant Genetic Resources, New Delhi-110012
Introduction
Some of the major challenges for the worldwide okra production include quick evolution and recent spread of begomoviruses, the emergence of novel viral strains and increasing abundance of viruliferous whitefly vectors (Venkataravanappa et al., 2015). Both conventional and non-conventional or genomic approaches are being applied for the management of this menace which is thoroughly discussed in this section. In the conventional approaches, the essential prerequisite for improving disease resistance is the availability of a suitable source of resistance (Dhankhar et al., 2005). Alternative sources of resistance to these viral diseases may be available in wild species like A. manihot, A. crinitus, A. angulosus, including certain landraces of A. tetraphyllus (Singh et al., 2007). But, for the successful transfer of desirable resistant genes from the wild-species to the A. esculentus, the botanical distance act as a barrier. Alternatively, all available germplasm should be evaluated under natural and artificial epiphytotic conditions to address the immediate problem of viral diseases. Some success has been attained in transferring Yellow vein mosaic virus (YVMV) resistance genes from the wild-species into cultivated okra species resistant cultivars such as P7 ( Abelmoschus esculentus × A. manihot ssp. manihot), Parbhani Kranti (A. esculentus × A. manihot), selections 4 and 10 (A. esculentus × A. manihots sp. tetraphyllus ) (Jambhale and Nerkar, 1986) have been released. Although, more than 50 improved varieties and hybrids have been released in India, but resistance to begomoviruses is found unstable with frequent breakdowns. This may be due to the pathogenic variability, selection of okra lines which are symptomless carriers or due to the emergence of polyphagous “B” biotype of Bemisia tabaci with its increased host-range. An alternative approach to controlling YVMV and Okra enation leaf curl virus (OELCV) incidence in okra are through the reduction of whitefly vector populations using insecticides, physical barriers, or by removal of symptomatic virus-infected plants. However, whiteflies are showing moderate to strong resistance to various insecticides. Therefore, it is required to adopt nonconventional methods of breeding, combining biotechnological tools for the development of pre-breeding lines resistant to viral stresses (Sanwal et al., 2014).
Management: Use of novel biotechnological tools will help in the achieving resistance against begomovirus in okra. Identification of markers linked to the resistance genes and its pyramiding for combining multiple disease resistance genes in various backgrounds should be done. The advancements in recombinant DNA technology and the optimization of transformation and regeneration protocols in okra has enabled the use of a genetic engineering approach for breeding. The pathogen-derived resistance (PDR) strategy seems very promising for the management of begomoviruses since it involves engineering resistance by transforming a susceptible plant using pathogen-derived gene sequences. The PDR can be achieved through various ways such as capsid protein mediated resistance, movement protein-mediated resistance, defective interfering (DI) viral DNA, incorporation of genes in antisense orientation, and truncated or mutated replicase (Rep) (Cl or ACl) gene (Lapidot and Friedmann, 2002). A transgenic is already developed for the okra shoot, and fruit borer (Narendran et al., 2013); thus, it is required to develop more transgenics especially using Coat protein (CP) genes for viral diseases like YVMV and ELCV. However, okra still lacks very effecient and robust transformation protocol. In addition, viral sequences shared by several begomoviruses can be exploited for the development of pathogen-derived resistance in okra (Lapidot and Friedmann, 2002). Also, introgressed resistance genes from wild okra species may complement the transgenic plants showing only partial resistance. Although, Venkataravanappa and Reddy (2013) have shown the use of non-radioactive DNA probe for the routine large-scale diagnosis of geminiviruses affecting okra. But, cost-effective and user-friendly DNA-based diagnostics should be developed for the large-scale screening of the germplasms for viral disease resistance.
Biotechnological strategies for improving viral disease resistance in okra
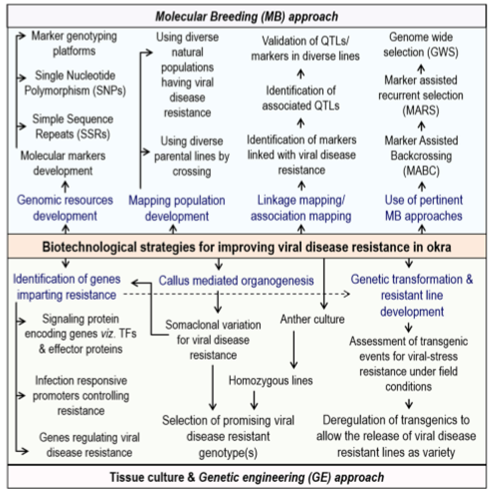
Limitations: Although RNA sequencing is relatively cheap, but limited funds available for research on genomically orphan crops such as okra, restrains the ability for deep sequencing of the transcriptome, leading to short unigenes representing only partial gene sequences. Therefore, deep-sequencing will result in long unigenes, representing full length gene sequence, which will be more useful for further biotechnological studies. The absence of an effecient transformation system has hampered the progress in okra genetic engineering research. Regarding in vitro culture of okra, only limited numbers of protocols were reported for shoot organogenesis.
Conclusion: Despite the importance of okra, as one of the important vegetable crop, very little attention has been paid to its genetic improvement using advanced biotechnological tools. The exploitation of marker assisted breeding in okra is often limited due to the availability of a few molecular markers, the absence of molecular genetic-map(s), and other molecular tools. However, high throughput biotechnological tools like chromosome engineering, RNA interference (RNAi), marker-assisted recurrent selection (MARS), genome-wide selection (GWS), targeted gene replacement, next generation sequencing (NGS), and nanobiotechnology can provide a rapid way for okra improvement.
References:
Venkataravanappa, V., Reddy, C. N. L., Jalali, S., Briddon, R. W., and Reddy, M. K. (2015). Molecular identiï¬cation and biological characterization of a begomovirus associated with okra enation leaf curl disease in India.Eur.J.Plant Pathol. 141, 217-235. doi: 10.1007/s10658-014-0463-0
Dhankhar, S. K., Dhankhar, B. S., and Yadava, R. K. (2005). Inheritance of resistance to yellow vein mosaic virus in an interspeciï¬c cross of okra (Abelmoschus esculentus). Indian J. Agr. Sci. 75, 87-89.
Singh, B., Rai, M., Kalloo, G., Satpathy, S., and Pandey, K. K. (2007). Wild taxa of okra (Abelmoschus species): reservoir of genes for resistance to biotic stresses. Acta Hortic. 752, 323-328. doi: 10.17660/ActaHortic.2007.752.55
Jambhale, N. D., and Nerkar, Y. S. (1986). �"Parbhani Kranti", a yellow vein mosaic resistant okra. Hortic. Sci. 21, 1470-1471.
Sanwal, S. K., Singh, M., Singh, B., and Naik, P. S. (2014). Resistance to yellow vein mosaic virus and okra enation leaf curl virus: challenges and future strategies. Curr. Sci. 106, 470-1471.
Lapidot, M., and Friedmann, M. (2002). Breeding for resistance to whitefly-transmitted geminiviruses. Ann. Appl. Biol. 140, 109-127. doi: 10.1111/j.1744-7348.2002.tb00163.x
Narendran, M., Shirale, D., Parimi, S., Deole, S. G., Nanote, A., Char, B. R., et al. (2013). Effecient genetic transformation of okra (Abelmoschus esculentus (L.) Moench) and generation of insect-resistant transgenic plants expressing the cry1Ac gene. Plant Cell Rep. 32, 1191-1198. doi: 10.1007/s00299-0131415-4
Venkataravanappa, V., and Reddy, M. K. (2013). Begomovirus characterization, and development of phenotypic and DNA-based diagnostics for screening of okra genotype resistance against Bhendi yellow vein mosaic virus. 3Biotech 3, 461-470. doi: 10.1007/s13205-012-0107-z
About Author / Additional Info: